Logbook¶
Day 1 (6/14): Completed the Python tutorial using carpentries. Learned how to create/utilize variables, process tabular data files, visualize tabular data files, store values in lists, repeat actions using loops, analyze data from multiple files, and debug programs.
Day 2 (6/15): Completed the Version Control with Git tutorials using carpentries. Learned how to create a repository in Git, record changes in Git, read tabular data, modify and commit files, utilize the HEAD and CAT commands, and push/pull/clone a remote repository.
Day 3 (6/16): Completed the Unix tutorial using carpentries. Learned how to create files and directories, navigate files and directories, construct command pipelines, process files, write loops and shell sripts, and utilize the commands grep and find.
Day 4 (6/17): Read "The iPlant Collaborative: Cyberinfrastructure for Enabling Data to Discovery for the Life Sciences" and "Ten Simple Rules to Cultivate Transdisciplinary Collaboration in Data Science". Learned about the origins of CyVerse and how it was initially used to support life science communities in their search for a proper tool for data management. Understood the importance of following ten simple rules to help with furthering the culture of collaboration.
Day 5 (6/18): Met with Dr.Swetnam and discussed my tasks for the day.Created this website in order to track progress during KEYS and comply with reproducible science. Added hyperlinks to the introduction page and logbook, created each of the pages using GitHub, and added descriptions to the introduction and logbook pages.
Day 6 (6/21): Added descriptions and hyperlinks to the CyVerse, R, and Python pages. Completed my logbook with more detail. Learned how to launch GitPod and then opened a Jupyter Notebook using GitPod.
Launching GitPod for the first time!
Launching Jupyter Notebook using GitPod
Day 7(6/22): With the help of Dr.Swetnam learned how to access Docker from GitPod and run GodLoveD/lolcow container using Docker.
Output from my first container on GitPod
Manipulating the various entry points of the container using GitPod
Day 8(6/23): Attended the KEYS Science Literacy Seminar in the morning where the deans of several of the colleges at the University of Arizona talked. During the seminar also discussed the need to present our projects in layman terms so that the average person can understand what is happening. In the afternoon met with Dr.Swetnam and discussed the tasks for the upcoming two days. Tried to build container using GodLoveD/lolcow as an example, but came across this error.
Error while attempting to build and push first container
Day 9(6/24) Attempted to launch Rstudio using the GitPod extension, but was unable to due to errors. Researched about these errors on the GitPod community, but was not able to find any solutions. Used KasperBrink's GitHub repository as an outline for this attempt. Created pages on the website for Docker, GitPod, GitHub Actions, and Java. Added information + pictures to the Docker page.
Error while attempting to launch RStudio using GitPod
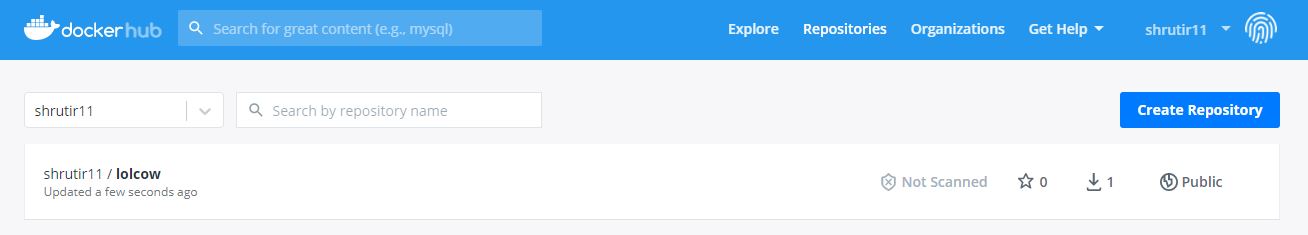
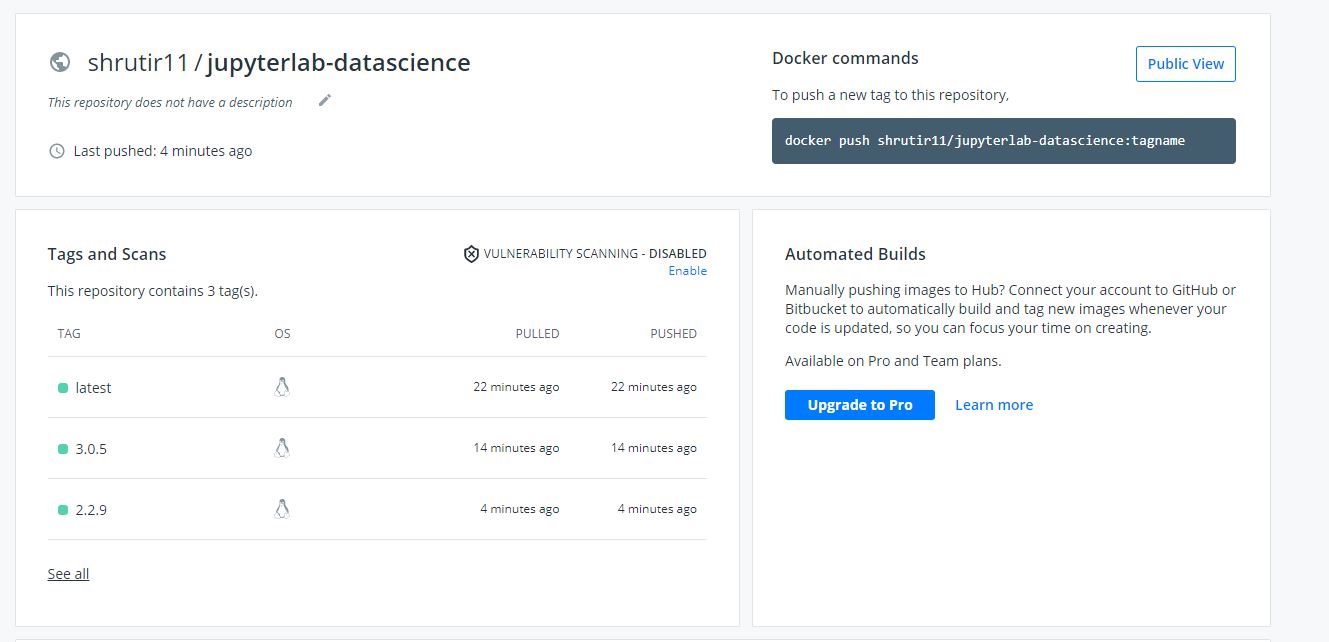
Day 10(6/25) Added descriptions and pictures to the GitPod, GitHub Actions, and Java pages. With the help of Dr.Swetnam was able to build and push first container to DockerHub.
Pushing first container to DockerHub
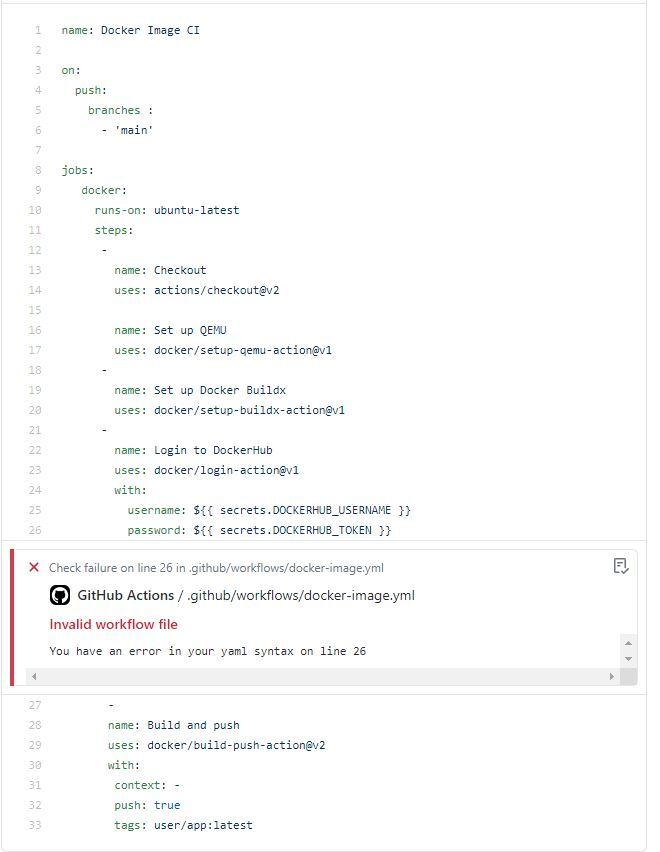
Day 11(6/28) Added the materials and methods page to the website and added descriptions to these pages. Tried to push container from GitHub to DockerHub. Faced several problems along the way two of the problems are shown below.
Error while pushing container to DockerHub/ Solved the error by creating an access token through DockerHub
Error while attempting to build the container
Day 12(6/29) Met with Dr.Swetnam and he recommended I try creating a seperate repository to try and push a container to DockerHub to see if that works first before attempting this with the rstudio-verse repository. Created a seperate repository using a tutorial and was able to push my first container from GitHub to Docker using GitHub Actions. Hope to replicate this in the upcoming days for the rstudio-verse repository.
First checkmark in GitHub Actions indicating my container was pushed to DockerHub!! Took probably over 25-30 tries
Container appearing in DockerHub after it was pushed from GitHub using GitHub Actions
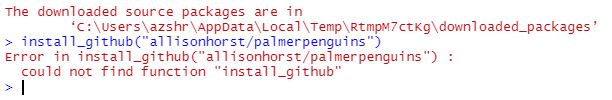
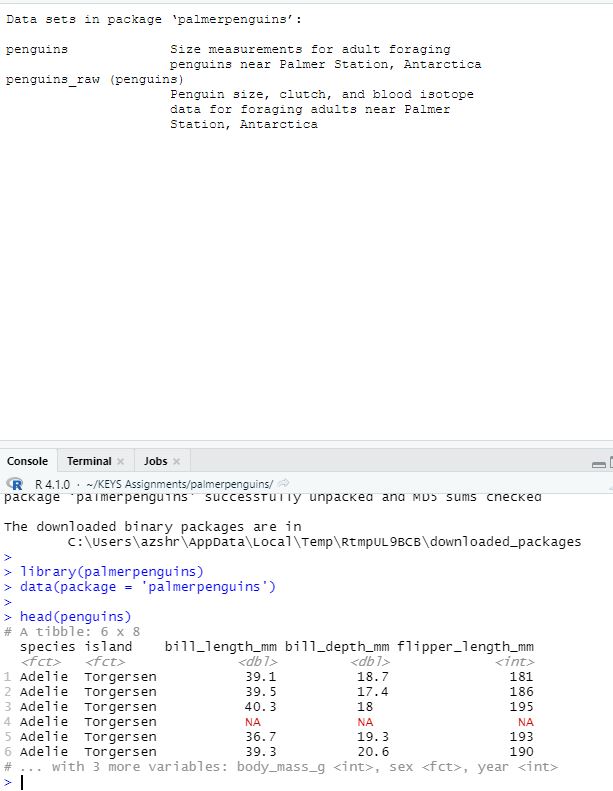
Day 14(7/6) Opened new project in RStudio and played around with the dataset PalmerPenguins. Followed the instructions to explore the dataset on this website Had trouble at first installing package in the RStudio library. Solved the problem with the help of Dr. Swetnam and this article
Error when trying to install palmer penguins package in RStudio
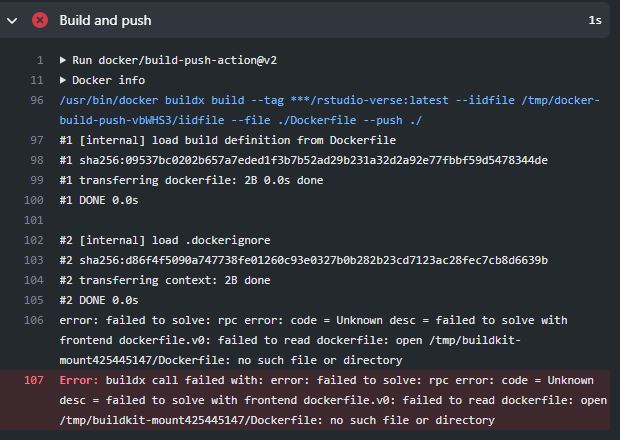
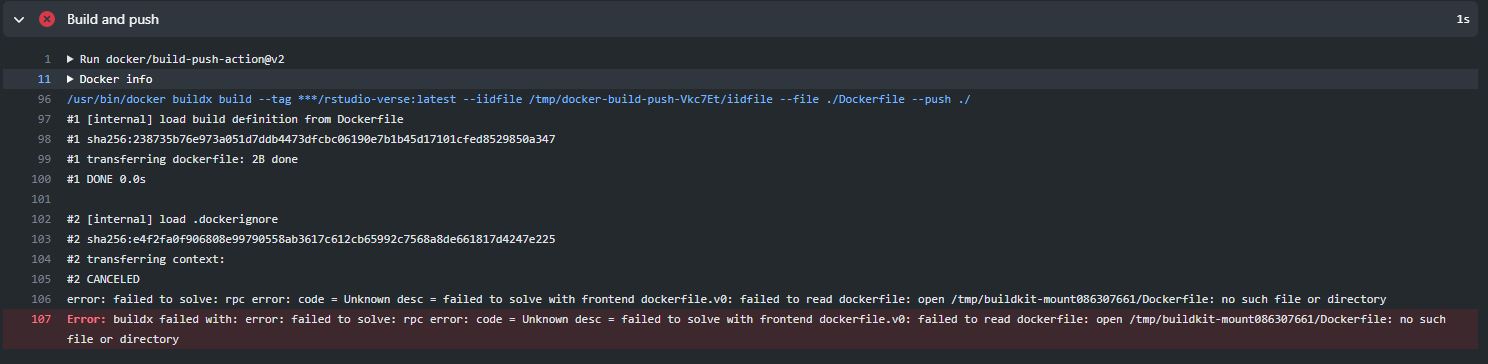
Palmer Peguins package was successfully installedDay 15(7/7) Tried to push rstudio-verse container to Docker using GitHub Actions. Faced several problems along the way. One being that during the build and push stage GitHub was unable to locate the Dockerfile since there were no Dockerfiles in the root of the rstudio-verse folder. To solve this problem created a path and identified where the Dockerfile was. Took about 50+ tries to do this. Then faced another problem when GitHub was installing the rockerverse container because the code exited at step 14/26.
During Build and Push unable to locate Dockerfile
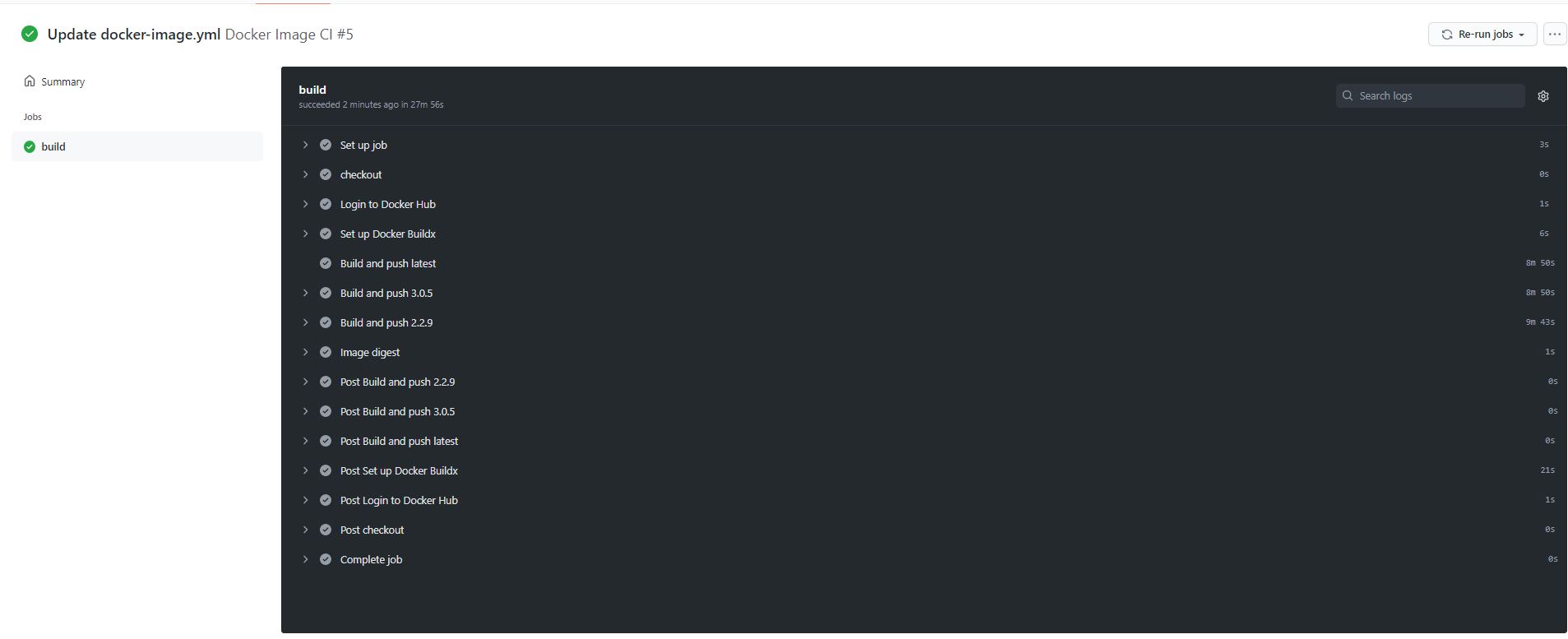
Day 16(⅞) The most successful day of this internship!! Launched RStudio-verse:latest container to DockerHub. Proceeded to use GitHub Actions and launched the rest of the versions of rstudio-verse to DockerHub. Still facing problems with one of the versions of RStudio-verse(3.6.3). Think the problem might be in the Dockerfile and not the .yml file because the code exits at a certain step in the Dockerfile. Was also able to launch all three versions of JuptyerLab to Docker using GitHub Actions under one .yml file.
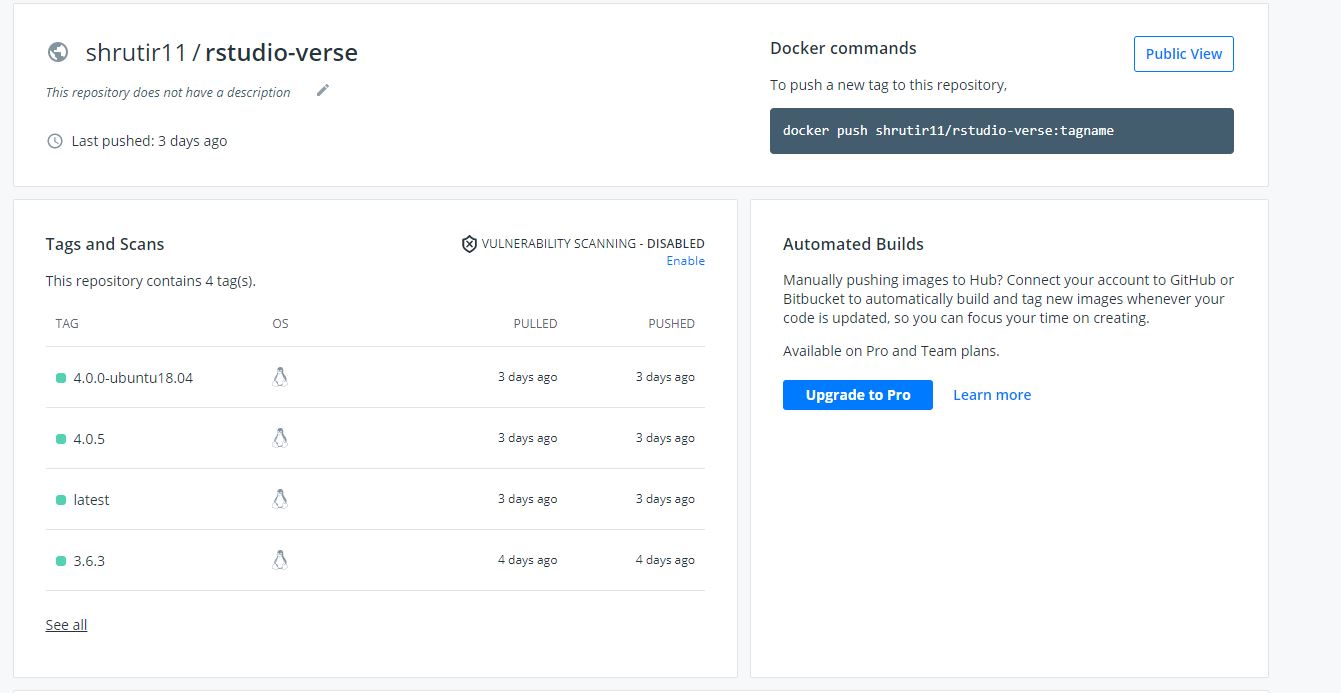
All four versions of RStudio pushed to DockerHub
Successful build and push of three versions of JupyterLab using GitHub Actions
All three tagged versions of JupyterLab pushed to Docker using one .yml file
Day 17(7/9) Read this page and learned how to schedule automatic workflow builds. Tested this by setting a time on Friday and seeing if it worked. It worked. Then scheduled the actual automatic build to take place every Saturday at 12:00 UTC time.
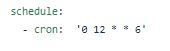
Code to schedule automatic workflow build
Day 18(7/12) Scheduled builds failed over the weekend so attempted to fix those. Started and finshed the conclusion and discussion of results assignment
Day 19(7/13) Started to work on poster and presentation. Added problem, solution, and materials to both poster and presentation. Created graphic for methods on biorender.
Day 20(7/14) Finalized poster and presentation before sending to Dr.Swetnam for feedback. Received feedback and changed the poster and presentation accordingly. Wrote short abstract and received feedback from Dr. Swetnam.